
Metabolomics
for Maximizing Research Impact
Why General Metabolics
GMet is different from other metabolomics providers. The company was founded to eliminate barriers to the use of metabolomics in life sciences research. We bring speed, rigor, efficiency, and biological insight to metabolomics scientists.
GMet was created by academic and industrial leaders in metabolomics with a strong track-record of harnessing its utility in life sciences. We bring our extensive metabolomics experience to each client’s project.
Our unique process efficiency drives cost efficiency for your research. GMet provides highly competitive pricing across all its services, enabling studies that would be prohibited by cost or duration by other platforms.
We use streamlined workflows to measure a wide range of metabolite classes. Our high throughput standardized analytical methods require very small sample volumes and eliminate most of the need for time-consuming, bespoke method development.
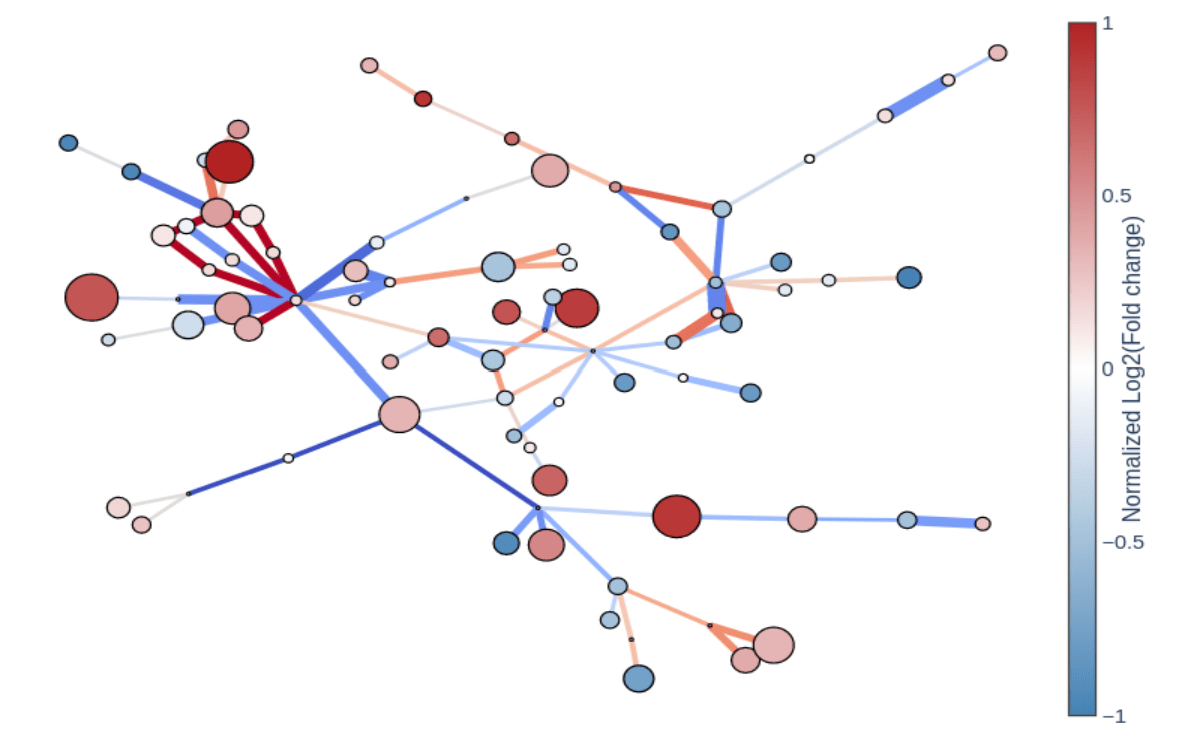
Our metabolomics analysis software bridges the needs of data scientists, biologists, and analytical chemists, providing QC data, differential analysis, cohort statistics, pathway maps, pathway enrichment tools and multi-omics tools in an intuitive, interactive environment where biologists can meet their partners in data science for optimal collaboration.
Our goal is to improve access to metabolomics for all researchers, enabling it to function at the speed and scale of the other tools of life science, and ensure you get the most out of your metabolism data. We work directly with you to plan your experiment, make sure you are acquiring the right data with the right tool, and help you interpret your results with the maximum speed to insight.
What is Metabolomics?
Metabolomics is the primary tool used to understand variation in metabolism. It refers to the simultaneous measurement of hundreds to thousands of small molecule metabolites in a cell, tissue, or bio-fluid.
Discuss Your Project Today
With the Experts in Metabolism

WHAT OUR CLIENTS SAY
Discuss Your Project Today
With the Experts in Metabolism Research
GMet is a unique metabolomics service provider founded by leaders in the field of metabolomics with a strong track-record of harnessing its utility in life sciences. Contact us today to discuss your project with us.