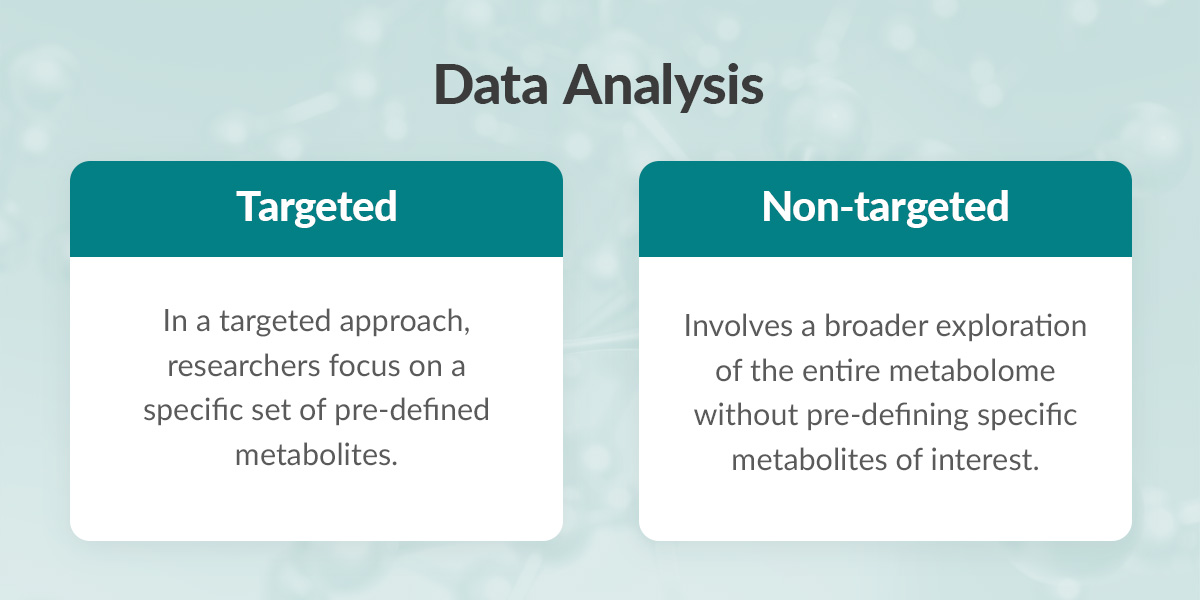
Metabolomics Defined
Individual metabolites can provide very focused information about biology: a particular mutation, genotype or disease state can be characterized by aberrant levels of specific metabolites (e.g. elevated triglycerides linked with cardiovascular disease). Metabolomics goes beyond individual metabolite assays, delivering metabolite level profiles in a comprehensive picture of very broad sets of all the small molecule metabolites within a biological system. These metabolites encompass many natural compound classes, such as sugars, sugar-phosphates and carboxylic acids; amino acids, fatty acids and lipids; nucleotides, and small molecule hormones, as well as many other organic molecules.
To capture this idea of the full complement of small molecules in biology, the term “metabolome” first surfaced in 1998 in a review article by scientist Stephen Oliver. Over the two decades since the term’s inception, pioneers in the field have conducted extensive research to inform and shape this area of science… including the team of scientists behind General Metabolics, who have been deeply engaged in the development of metabolomics since the inception of the field.
Today, the analysis of metabolites has expanded to include comprehensive analysis of hydrophobic molecules called fatty acids and lipids – frequently referred to as “lipidomics.” With this distinction, metabolomics is now more often thought of as delivering data on the more polar and hydrophilic metabolites, while lipidomics provides insight into hydrophobic classes of metabolites such as phospholipids, steroids, and signaling molecules such as eicosanoids and prosteglandins. There are not clear lines that define which species are polar metabolites vs. lipids.
The scope of metabolomics is broad. Some key areas where metabolomics is vital include biomarker discovery, disease research, precision medicine, environmental studies and nutritional sciences.