- More efficient to acquire, process, and interpret
- More accessible to data scientists, analytical chemists, and biologists,
- Easier to connect to and integrate with other data types (e.g. metadata or other omics data).
Platform Overview, Platform Architecture, Tools for Lab Operations, Tools for Researchers
The elements of our metabolomics platform enable rapid turnaround, alleviate computational intensity, and drive researchers rapidly to biological insight. We couple a full suite of LC-MS tools for data acquisition with an intuitive informatics ecosystem to process, analyze, and interpret metabolomics and lipidomics to support life sciences research.
GMet’s Interactive data reporting and analysis tools enable researchers to focus on biologic hypothesis generation and data exploration. GMet software can also serve as a bespoke laboratory information management system (LIMS) to enable coordination across many experiments.
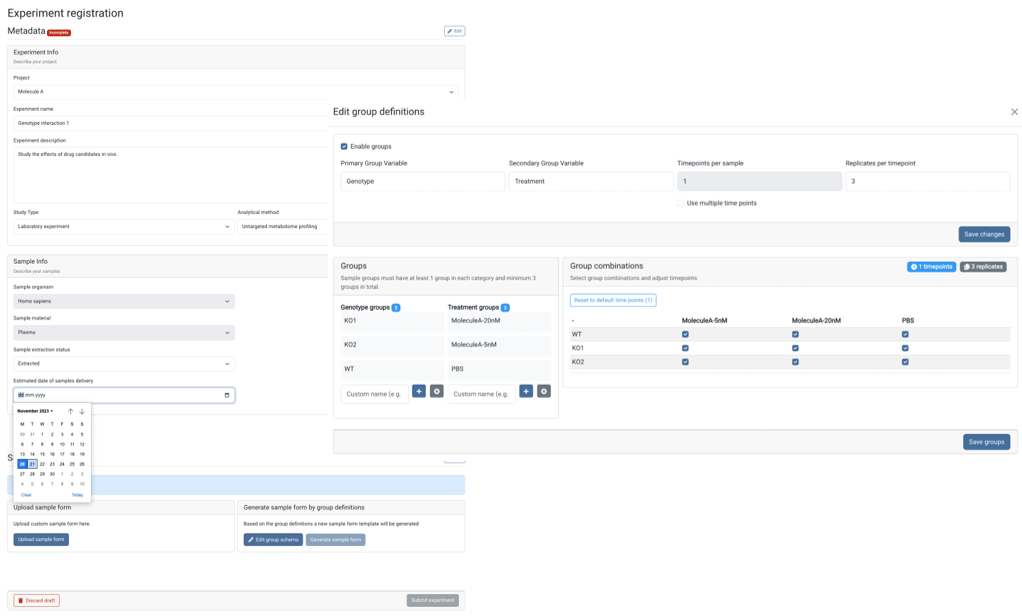
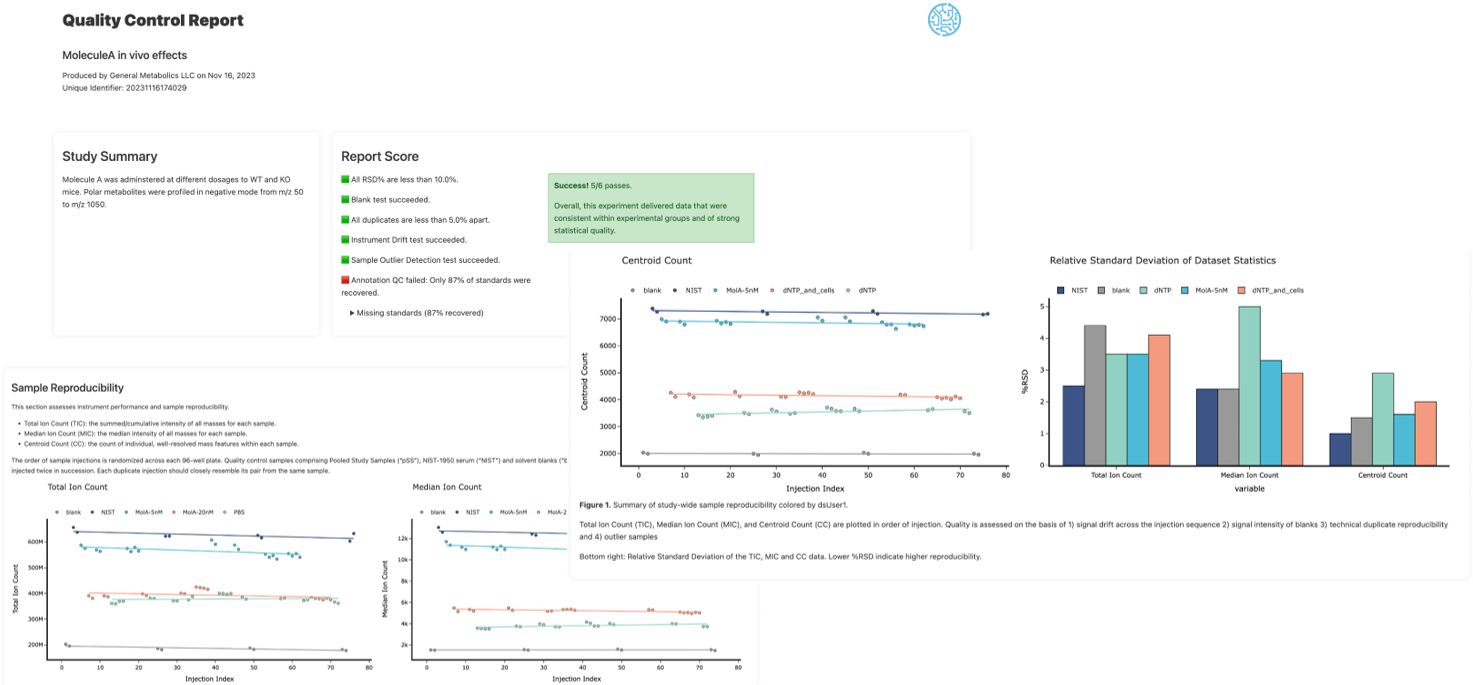
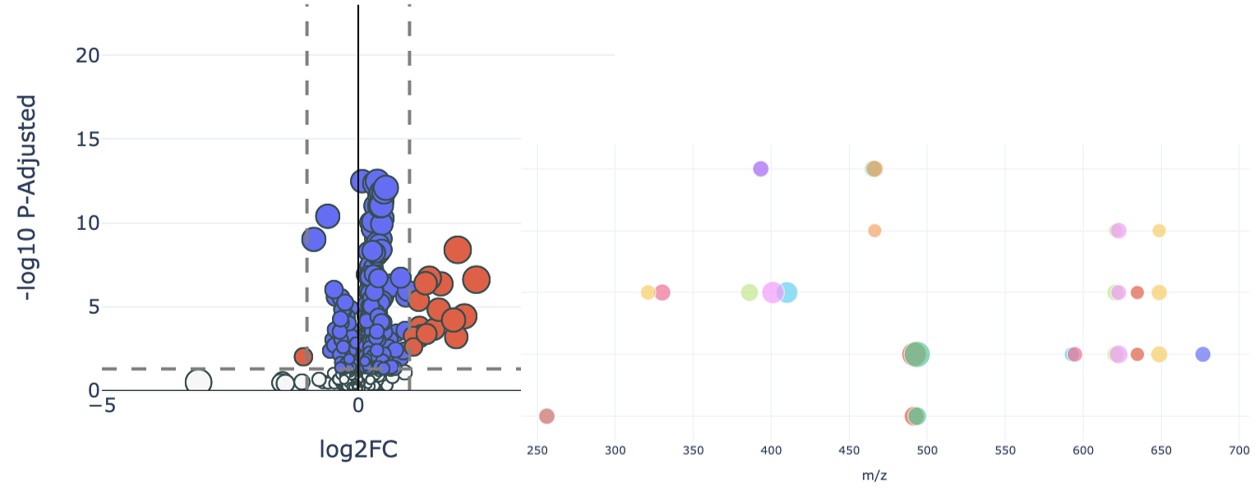
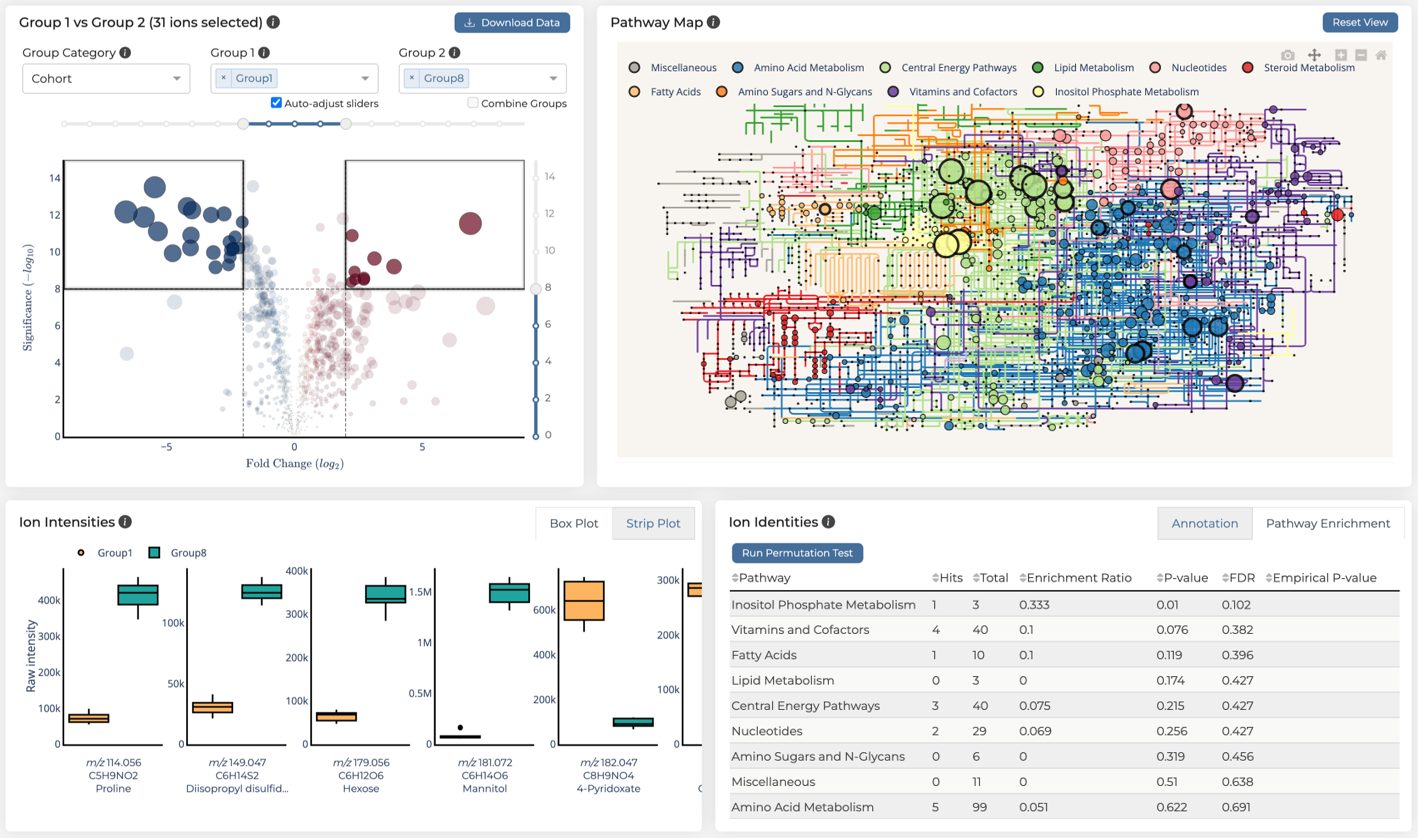
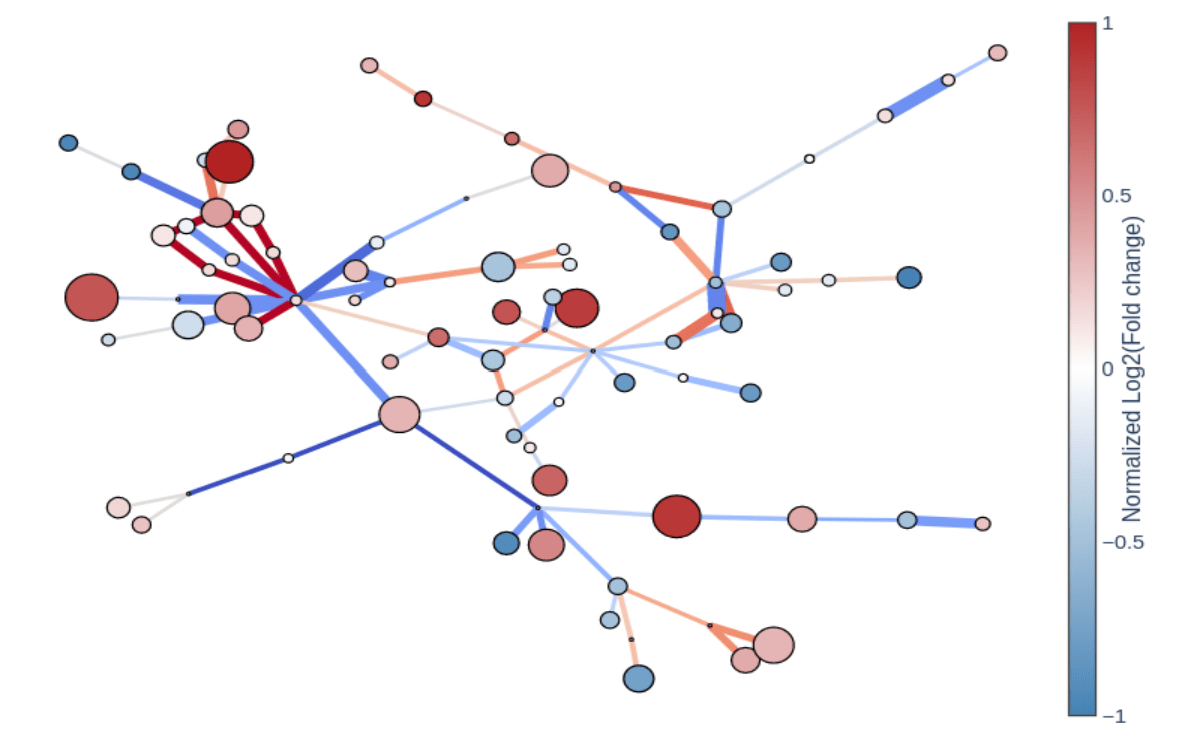
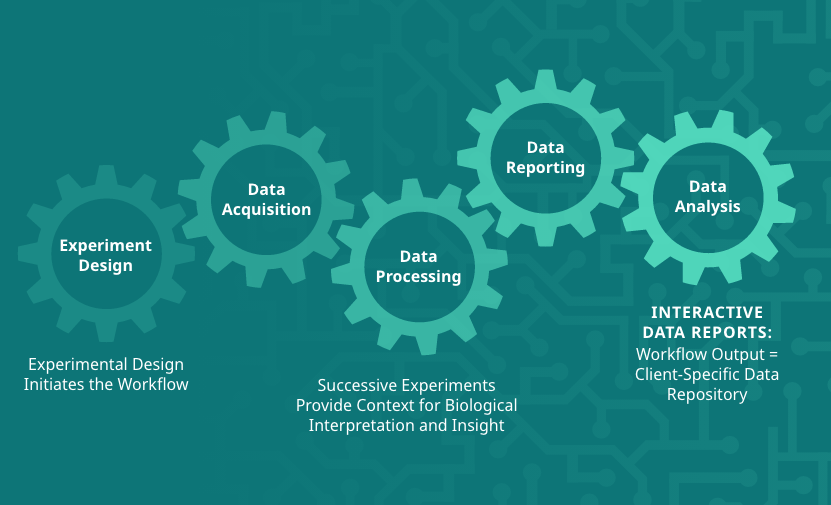
The GMet metabolomics platform is designed to streamline the acquisition, processing, analysis and interpretation of metabolomics data. Individual components of the platform are linked in an integrated data pipeline to efficiently handle the unique data storage, management, and analysis challenges presented by metabolomics data. This enables us to efficiently support a wide range of study scales, from smaller lab-based experiments up to larger population scale studies of many thousands of samples.
Our platform provides software to capture experimental design and meta-data, utilizes high throughput methods for data acquisition, employs sophisticated algorithms to automate data processing and combines these in an integrated data pipeline to automate reporting, output generation, and data visualization. Interactive data analysis tools complement GMet’s metabolomics analysis software to facilitate biological interpretation and hypothesis generation.
GMet Software contains tools to support efficient lab operations that can be deployed anywhere through the cloud including, experimental design & meta data capture, integrated tooling for instrument configuration and data processing, parallelized cloud native data processing workflows, and automated quality control (QC) reporting.
